gaussdev (replaces TGaussDev)
The gaussdev function returns a 1-dimensional array of random numbers distributed according to a Gaussian probability density function. These values are known as "Gaussian Deviates". The larger the number of values created, the more perfectly they will approximate a Gaussian normal distribution.
tbl = gaussdev( nCount )
tbl = gaussdev( nCount, nStdDev )
tbl = gaussdev( nCount, nStdDev, nMean )
![]() nCount is the number of Gaussian Deviates to
create.
nCount is the number of Gaussian Deviates to
create.
![]() nStdDev is
the standard deviation. If omitted, it is set to 1.0.
nStdDev is
the standard deviation. If omitted, it is set to 1.0.
![]() nMean is
the mean value. If omitted, it is set to 0.0.
nMean is
the mean value. If omitted, it is set to 0.0.
![]() tbl is the returned array containing
nCount
values.
tbl is the returned array containing
nCount
values.
![]() On
failure, nil is
returned.
On
failure, nil is
returned.
The following script creates 1 million Gaussian random deviates, creates a histogram of the values, and plots the histogram. The random values are not symmetrical about the middle since they are drawn from a random population. To make this script brief, the default histogram binning option, "Auto", is used. The auto method computes the minimum and maximum of the values created and selects the number of bins betwee, Since these values are not specified, the script retrieves the histogram binning values actually used and then uses them for making the plot.
|
|
-- create 1 million random deviates |
|
|
-- create a Histogram object |
|
|
-- compute the histogram |
|
|
-- separate the 3 parameters for clarity |
|
|
|
|
|
|
|
|
-- create a series for the histgram x axis |
|
|
-- plot the histogram |
|
|
|
Below is the result of running this script. The plot uses default labels as they were not specified in this plot function call.
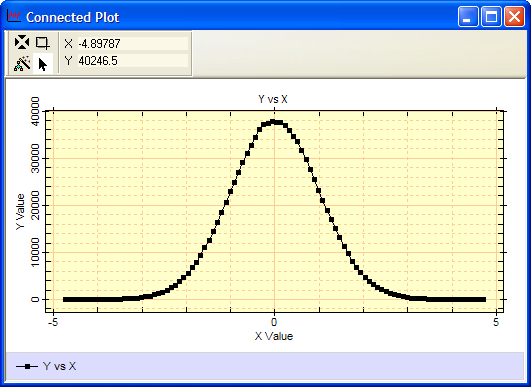
Table and Array Functions, random, series, CStats class
Mira Pro x64 Script User's Guide, Copyright Ⓒ 2023 Mirametrics,
Inc. All Rights Reserved.